Leave a comment at the end of this page or email contact@krishagni.com
Create EasyBlood aliquots
Prerequisites
- This import creates a new box if not present (Target_RackBarCode field value — Assigned to Box Unique Name).
- If the box with the same name exists, it will be reused. Import will give an error if the box has specimens in the same position as in the CSV.
- The newly created boxes are stored in a temporary dimensionless freezer.
Note: The dimensions of newly created boxes are fixed to 8 X 12 with alpha-numeric Labeling Scheme. This is not configurable. - Please refer Dimensionless Containers wiki page to create a dimensionless freezer.
- After creating the freezer, use the below steps to configure
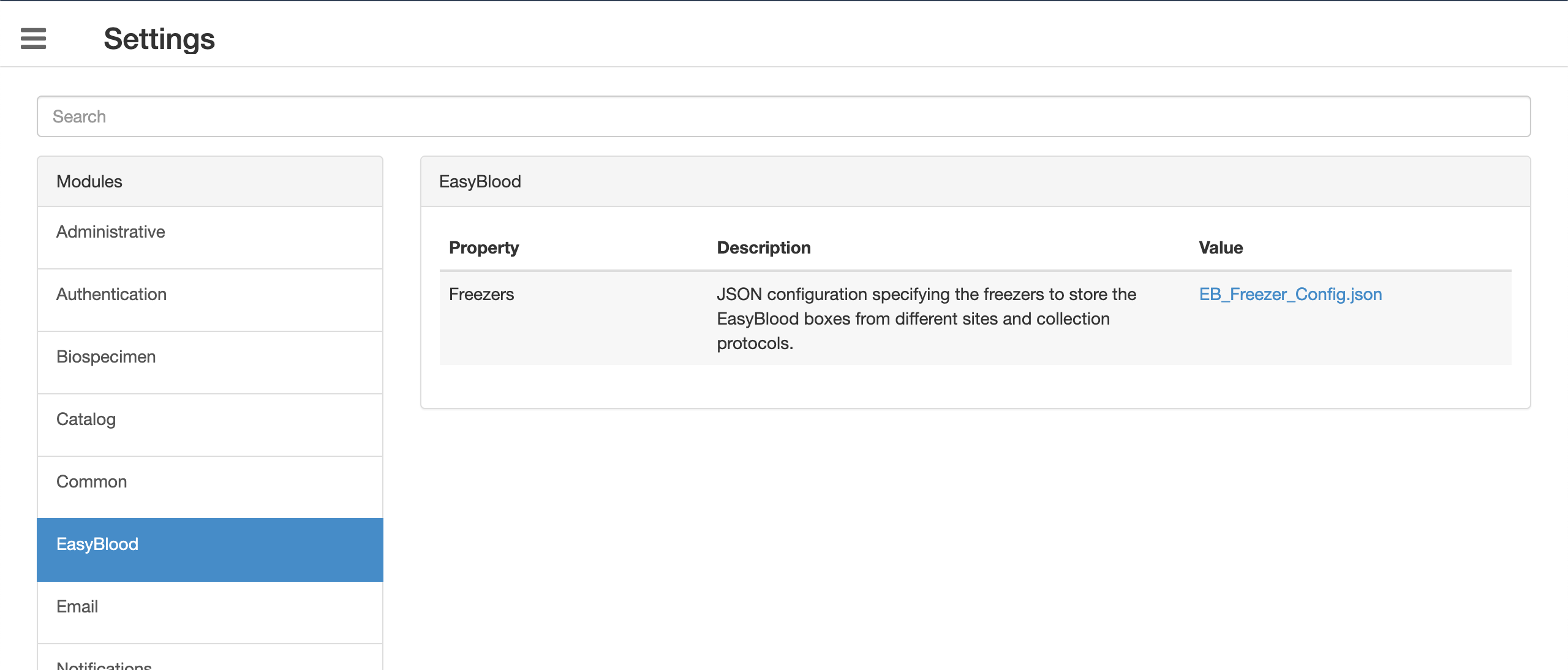
- Go to Settings and search for easyBlood
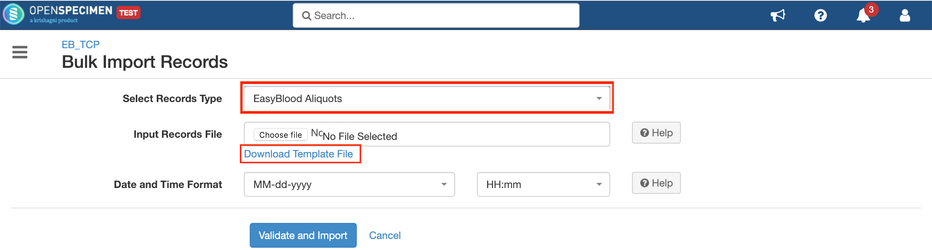
Download sample working JSON
Here, '<site_prefix>' maps to the column 'EB Code' in the input CSV.
- Go to Settings and search for easyBlood
- The importer will show an error 'Site or parent container detail is required' if the freezer is not specified for the corresponding site or CP.
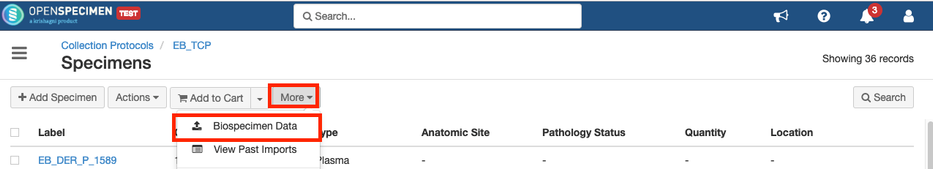
Steps to import CSV
Sample CSV
Data Dictionary
| Column Name | Datatype | Mandatory? | Description | Permissible Values | Validations |
|---|---|---|---|---|---|
| ActionDateTime | Date | No | This is taken as the aliquot creation date. It will take the current date value if nothing specified. | This date should not be less than the parent specimen | |
| Fraction Type | String | Yes | The specimen type | Specimen-type-permissible-values.csv | |
| Source_Barcode | String | Yes | The label for the parent specimen | The parent specimen must exist | |
| Target_Barcode | String | Yes | The label for the aliquot specimen | ||
| Target_Volume | Integer | No | The quantity for aliquot specimen | ||
| ErrorCode | Integer | No | Error information about the aliquot while processing from EasyBlood | ||
| ErrorText | String | No | Error information about the aliquot while processing from EasyBlood | ||
| Target_RackBarCode | String | Yes | The target freezer container name | This container should exist in OpenSpecimen. If not, the box would be created under the intermediate freezer. | |
| Target_PositionID | String | Yes | The position ID is (row, column). The first character identifies the row. The row is specified using an English letter (A - Z). The rest of the characters in position ID identify the column. The column is a number. D50 identifies the 50th column of row D. The target container position, ex. A1 (First row, first Column), D5 (Fourth row, fifth column) | {A,B, .., H} {1,2,3, .., 12} | No other specimen should be present in this position. |
| EB Code | String | No | This is the site_prefix. When creating an intermediate freezer the appropriate freezer under this site_prefix would be computed from the JSON config file. | The cp and freezers mentioned in the JSON should be present in OpenSpecimen. | |
| Close Ancestors | boolean | No | This controls whether the ancestors of the aliquot being created, should be closed or not. Note: This will close all the parent specimens up to the primary (lineage='New') level if at least one aliquot has 'Close Ancestors' = 'true' |
Related content
Leave a comment at the end of this page or email contact@krishagni.com