In OpenSpecimen, we have features like auto-allocation, data validation (edit checks), etc. Some of these are also supported in workflows.
Container Auto-allocation
You must choose between the two allocation strategies and set them at the protocol level to use auto allocation in workflows. Refer to the wiki page for more details.
Once the auto-allocation is set up at the CP level, you need to add the below code in your workflow JSON:
enableAutoContainerAllocation: true,
Below is a JS file for reference:

Group assigner
You can add a group assigner within your workflow for respective tasks.
Steps to add group assigner in your workflow...
Example 1: User scans a total number of 4 specimen tubes (1 is SST, and 3 are EDTA). EDTA tubes should go for ficoll isolation, and SST should go for clotting.

Code to be added within workflow JSON:
{
"type" : "group-assigner",
"name" : "task-assigner-1",
"title" : "Select Next Task",
"shortDescription" : "Assign tubes for next tasks",
"activityStatus" : "Active",
"longDescription" : "Points to consider when receiving specimens: <ul> <li> Send SST tubes for clotting </li> </ul><ul> <li> Send EDTA tubes for ficoll isolation </li> </ul><ul> <li> Send Citrate tubes for seperating out plasma and buffy coat </li> </ul>",
"jsonConfig" : false,
"starterTask" : false,
"optional" : false,
"inputs" : [ {
"sourceTask" : {
"type" : "update-specimens",
"name" : "receive-specimens",
"title" : "Receive Specimens",
"shortDescription" : "Receive the whole blood tubes that have to be processed",
"activityStatus" : "Active"
},
"sourceKey" : null
} ],
"userGroups" : [ ]
}
Below is the JS file:

Example 2: If the anatomic site of fresh tissue is the ‘Thyroid gland', it should go for the pool specimens step, and if the anatomic site is the 'Parathyroid gland’, it should go for the fixation step.

Code to be added within workflow JSON:
{
"type" : "group-assigner",
"name" : "task-assigner",
"title" : "Next Task Assigner",
"shortDescription" : "Assign next task to tissue samples",
"activityStatus" : "Active",
"longDescription" : "Decide if you want to pool the fresh tissue specimens and then process them further or directly start processing the samples as is.",
"jsonConfig" : false,
"starterTask" : false,
"optional" : false,
"inputs" : [ {
"sourceTask" : {
"type" : "update-specimens",
"name" : "receive-specimens",
"title" : "Obtain a fresh specimen",
"shortDescription" : "Obtaining fresh specimens from collection centers",
"activityStatus" : "Active"
},
"sourceKey" : null
} ],
"userGroups" : [ ]
}
Below is the JS file:

Skip Tasks
When defining your workflow, you configure it based on the standard procedure. However, there may be deviations for some batches of specimens where you do not perform specific tasks. In such cases, you will be able to skip those tasks.
Click here to expand...
When a task is skipped, you must specify its reason, and a ‘Deviation Event’ is added to the specimens.




Code | Description |
"onSkipAction" : "SKIP_DEPENDENTS" | The dependent tasks are skipped too. |
"onSkipAction" : "COPY_INPUT" | The input of the last task is copied over to the next task. |
Below is the example code:
{
"type" : "form-data-entry",
"name" : "clotting",
"title" : "Rest period for blood tube",
"shortDescription" : "60 minutes after collection",
"activityStatus" : "Active",
"longDescription" : "Rest period for blood tube: <ul> <li> 60 minutes after collection, in upright position </li> <li> Register date and time </li></ul>",
"jsonConfig" : true,
"configuration" : {
"formType" : "SpecimenEvent",
"formName" : "SpecimenClottingEvent",
"defaultValues" : {
"rest_time_in_minutes" : 60,
"comments" : "N/A"
},
"fields" : [ "rest_time_in_minutes", "time", "comments" ]
},
"starterTask" : false,
"optional" : true,
"onSkipAction" : "SKIP_DEPENDENTS",
"inputs" : [ {
"sourceTask" : {
"type" : "group-assigner",
"name" : "task-assigner-1",
"title" : "Select Next Task",
"shortDescription" : "Assign tubes for next tasks",
"activityStatus" : "Active"
},
"sourceKey" : null
} ],
"userGroups" : [ {
"id" : 143,
"name" : "Divya Team1",
"description" : "Divya Team1 to be used for workflows",
"institute" : "Institute Divya",
"activityStatus" : "Active"
} ]
}
Remove Specimens
Once you start a batch, there are chances that you want to remove one or more specimens from the batch due to any of the reasons stated below:
One of the specimens got damaged as it fell on the ground, and the tube broke.
The quantity received was insufficient.
The blood tube had blood clots.
The tube received was already frozen.
An incorrect specimen was selected from the waiting queue and added to the batch.
Skip Mode
You can remove the specimens from a batch/task and specify the reason for it. A deviation event will be added to that specimen.
NOTE: The skip mode will behave differently based on the type of task, as stated below:
Task Type | Behavior |
|---|
"type" : "update-specimens" | You will get an option asking if you want to ‘Skip only this task' or 'Skip all remaining tasks’. You can select one and specify the reason. |
"type" : "create-derivatives | It will not give you an option and will only ask for a reason for removing the specimens. |
"type" : "form-data-entry" | You will get an option asking if you want to ‘Skip only this task' or 'Skip all remaining tasks’. You can select one and specify the reason. |
"type" : "create-pooled" | There is no option of remove specimens from this task. If you wish to skip the task, you can add the ‘Skip' button for this step. Refer to 'Skip Tasks’ section. |
"type" : "create-aliquots" | It will not give you an option and will only ask for a reason for removing the specimens. |
Skip only this task
Click here to expand...
If you want to remove the specimen only from a specific task, select the 'Skip only this task' option and specify the reason:


Skip all remaining tasks
Click here to expand...
If you intend to skip all the tasks for a specimen, select the 'Skip all remaining tasks' option and specify the reason:


Remove from batch (v10.0)
Click here to expand...
If you added a specimen accidentally to the batch, you can remove it. This specimen will now get added back to the waiting queue.

Dispose Specimen (v10.1)
Sometimes after receiving a specimen or after creating multiple aliquots, the tube may get damaged or broken by falling on the floor or spilling of the sample. As a result, you might want to remove the specimens from the batch and mark them as disposed/closed.
Click here to expand...
The 'Dispose Specimen' will be available in the 'Remove Specimen' popup. This will allow you to remove the specimens using the 'x' icon, add a reason for removing the specimen, and also dispose off the specimens on the same screen as shown below:


The ‘Dispose Specimen' checkbox will only appear if you select ‘Skip all remaining tasks’ in the 'Skip Mode.’

Calculations based on various parameters
We have supported calculations based on multiple parameters within the workflows' module to make the data entry easy and avoid errors. This section will walk you through multiple calculation examples.
Live Calculation
To get a total cell count, you must multiply it by the cell count per ml with the dilution factor. It becomes difficult to manually calculate it on a calculator and then enter it in OpenSpecimen, leading to errors.
To avoid manual data entry errors and save time, we have added this feature where the calculation is done based on two fields (default and custom fields; both are supported).
Click here to expand...
Below is a short video depicting the same, where the ‘Quantity' is automatically calculated based on the values you enter in the ‘Cells Counted (million)’ and 'Dilution Factor’ fields:
2022-07-11 14-23-24.mkvExample code for the configuration:

Serum aliquots count & quantity based on primary specimen quantity
Usually, the quantity and count of aliquots are standard based on the parent specimen quantity. For example, a whole blood specimen of 10ml collected in a serum separator vacutainer should produce 6 aliquots of 0.5ml each, and a 3.5ml should produce 2 aliquots of 0.5ml.
Click here to expand...
Below is the depiction of the same:
2022-07-11 15-42-53.mkvExample code for the configuration:

PBMC aliquots count & quantity based on parent quantity
The number of PBMC aliquots to be created, and the quantity can be calculated based on its parent (derivative quantity). The calculation should be done as below:
Parent Quantity (millions) | No. of PBMC aliquots | Quantity of aliquots |
|---|
128 | 9 aliquots → 8 + 1 | 8 aliquots of 10 million 1 aliquot of 48 million |
1000 | 23 aliquots → 8 + 15 | 8 aliquots of 10 million 15 aliquots of 50 million |
44.5 | 4 | 4 aliquots of 11.125 million |
Click here to expand...
Below is the depiction of the same:
2022-07-11 17-54-45.mkvExample code for the configuration:

Calculate Spun Time based on Received Time and Spun Duration
Click here to expand...
The spun event time can be calculated as:
Spun Date and Time = Received Date and Time + Duration


Below is the depiction of the same:
2022-07-11 18-09-10.mkv
Example code for the configuration:

Calculate Spun Time based on Clotting Time and Spun Duration
Click here to expand...
The spun event time can be calculated as:
Spun Date and Time = Clotting Date and Time + Duration


Below is the depiction of the same:
2022-07-12 18-02-02.mkvExample code for the configuration:

Edit Checks (Data Validation)
Data validation is used to enter high-quality data. This ensures that the user does not violate any data entry rules while entering the data. Refer to the wiki page for more details.

Click here to expand...
Use cases Part 1:
Specimen received quality should be acceptable.
Derived plasma specimens should be at least 10 ml.
Example Code 1:
{
"name" : "editChecks",
"view" : null,
"ctrl" : null,
"data" : {
"constraints" : [ {
"records" : [ "specimen" ],
"rules" : [ {
"when" : "#specimen.lineage == 'New' && #specimen.status == 'Collected'",
"expr" : "#specimen.receivedEvent.receivedQuality == 'Acceptable'",
"description" : "Specimen received quality should be acceptable.",
"type" : "SOFT"
}, {
"when" : "#specimen.lineage == 'Derived' && #specimen.type == 'Plasma' && #specimen.status == 'Collected'",
"expr" : "#specimen.initialQty >= 10",
"description" : "Plasma derived specimens should be at least 10 ml.",
"type" : "SOFT"
} ]
} ]
}
}
If you proceed without fixing the deviations, the system adds a deviation event for the specimens.
You need to specify the code within the CP workflow. No need to change anything in the workflow JSON file.
Below is the example CP workflow:
.json?version=1&modificationDate=1652928949783&cacheVersion=1&api=v2&viewType=fileMacro)
Use cases Part 2:
Centrifugation is performed on the parent specimen multiple times, and the conditions should be as follows:
1st Centrifugation: Duration should be 10, and gForce should be 2000
2nd Centrifugation: Duration should be 20, and gForce should be 1000
3rd Centrifugation: Duration should be 20, and gForce should be 250
Example Code 2:
{
"when" : "#collFns.isNotEmpty(#specimenForms['SpecimenSpunEvent$Array']) && #workflow != null && #workflow.shortCode == 'FLUSH_PBMC' && #task != null && #task.name == 'spin-1'",
"expr" : "#collFns.forEvery(#specimenForms['SpecimenSpunEvent$Array'], 'SpunEvent', \"#SpunEvent['duration'] == 10\")",
"description" : "Duration should be 10 minutes.",
"type" : "SOFT"
}, {
"when" : "#collFns.isNotEmpty(#specimenForms['SpecimenSpunEvent$Array']) && #workflow != null && #workflow.shortCode == 'FLUSH_PBMC' && #task != null && #task.name == 'spin-1'",
"expr" : "#collFns.forEvery(#specimenForms['SpecimenSpunEvent$Array'], 'SpunEvent', \"#SpunEvent['gForce'] == 2000\")",
"description" : "gForce should be 2000 g.",
"type" : "SOFT"
}, {
"when" : "#collFns.isNotEmpty(#specimenForms['SpecimenSpunEvent$Array']) && #workflow != null && #workflow.shortCode == 'FLUSH_PBMC' && #task != null && #task.name == 'spin-2'",
"expr" : "#collFns.forEvery(#specimenForms['SpecimenSpunEvent$Array'], 'SpunEvent', \"#SpunEvent['recordId'] != #$inputFormRecordId || #SpunEvent['duration'] == 20\")",
"description" : "Duration should be 20 minutes.",
"type" : "SOFT"
}, {
"when" : "#collFns.isNotEmpty(#specimenForms['SpecimenSpunEvent$Array']) && #workflow != null && #workflow.shortCode == 'FLUSH_PBMC' && #task != null && #task.name == 'spin-2'",
"expr" : "#collFns.forEvery(#specimenForms['SpecimenSpunEvent$Array'], 'SpunEvent', \"#SpunEvent['recordId'] != #$inputFormRecordId || #SpunEvent['gForce'] == 1000\")",
"description" : "gForce should be 1000 g.",
"type" : "SOFT"
}, {
"when" : "#collFns.isNotEmpty(#specimenForms['SpecimenSpunEvent$Array']) && #workflow != null && #workflow.shortCode == 'FLUSH_PBMC' && #task != null && #task.name == 'spin-3'",
"expr" : "#collFns.forEvery(#specimenForms['SpecimenSpunEvent$Array'], 'SpunEvent', \"#SpunEvent['recordId'] != #$inputFormRecordId || #SpunEvent['duration'] == 10\")",
"description" : "Duration should be 20 minutes.",
"type" : "SOFT"
}, {
"when" : "#collFns.isNotEmpty(#specimenForms['SpecimenSpunEvent$Array']) && #workflow != null && #workflow.shortCode == 'FLUSH_PBMC' && #task != null && #task.name == 'spin-3'",
"expr" : "#collFns.forEvery(#specimenForms['SpecimenSpunEvent$Array'], 'SpunEvent', \"#SpunEvent['recordId'] != #$inputFormRecordId || #SpunEvent['gForce'] == 250\")",
"description" : "gForce should be 250 g.",
"type" : "SOFT"
}
Pool Specimens Task
Pooling is the process of merging one or more specimens of the same patient or multiple patients into a single specimen. Refer to the wiki page for more details.
Steps to add pool specimens task in workflow...
There can be two use cases for pooling of specimens at labs:
Use case 1: If there are 4 tubes (A, B, C, D), they are pooled together, and the original tubes are discarded. For this use case, add the below code in your JS file:
allowSpecimensInMultiplePools: false, // One tube can only be used in one pool
closeSpecimensUsedInPooling: true, // Closes the original specimen after pooling
Use case 2: If there are 4 tubes (A, B, C, D), tubes A and B are pooled together, some part of B is pooled with tube C, some part of C is pooled with tube D, and so on. In this case, a specimen from a single tube is used in more than one pool. For this use case, add the below code in your JS file:
allowSpecimensInMultiplePools: true, // One tube can be used in multiple pools
closeSpecimensUsedInPooling: false, // Does not close the specimen after pooling
To add a pool specimens task in your workflow, add the below code in your workflow.json file:
{
"type" : "create-pooled",
"name" : "pool-specimens",
"title" : "Pool Specimens",
"shortDescription" : "Mix two or more specimens into a single pooled specimen",
"activityStatus" : "Active",
"jsonConfig" : false,
"starterTask" : false,
"optional" : true,
"onSkipAction" : "COPY_INPUT",
"inputs" : [ {
"sourceTask" : {
"type" : "update-specimens",
"name" : "receive-specimens",
"title" : "Receive Specimens",
"shortDescription" : "Receive the 4 heparin tubes that have to be processed",
"activityStatus" : "Active"
},
"sourceKey" : null
} ],
"userGroups" : [ {
"id" : 142,
"name" : "Utrecht Team2",
"description" : "Utrecht Team2 Users",
"institute" : "UMC Utrecht",
"activityStatus" : "Active"
} ]
}
Below is the JS file for this task:

Default specimens in pool step (v10.0)
Based on your lab requirements, you can default which specimens are to be pooled by configuring some rules. Manually selecting the specimens every time can introduce errors.
Use case 1: Four Na-Heparin tubes are collected per participant, always. In this task, you pool the first two tubes and last two tubes together.


Example JSON:

Use case 2: Two Na-Heparin tubes are collected per participant, always. In this task, you pool the tubes of the same participant together.


Example JSON:

Workflow Manifest
Manifests are PDF files that can be downloaded or printed while processing specimens or after specimen collection. Refer to the wiki page for more details.
Steps to configure workflows manifest...
Create an HTML template based on your requirements in terms of which fields you want to display, the width of the columns, font size, etc.
Navigate to 'Workflows'.
On the workflows list view page, click on the inverted arrow as shown below:



Example HTML template:

Example Workflow Manifest:

Use case: Within a batch, there can be specimens from multiple participants. Just having a PPID for each task is not helpful, as it is just a unique identifier that the lab users do not understand. Having PHI data like D.O.B and Participant’s Name on each task will help them in distinguishing which patient's specimen they are dealing with and help in the reduction of errors. To configure participant fields in your workflow, refer to the JavaScript files of the tasks on this wiki page (Example 6).
When you start a batch, all the specimens in the batch might have the same values for certain fields within the form or update tasks. For example, if you have four whole blood tubes that undergo a wash step, the dilution volume and the date and time for all tubes will be constant. In such cases, it is easier if you can do it in a form mode where you need to enter the data only once, which will then copy the same values on all specimens within the task.
Click here to expand...
Screenshot of the task in table mode:

Screenshot of the task in form mode:

To configure the form mode, you need to add one of the below codes in your JS files:
"allowViewToggle": true // If you want both form and table mode within a task so that the users can toggle between the two modes and use the one they are comfortable with.
"viewMode": "form" // If you want to fix the form view by default for a task.
By default, only editable fields are displayed in the form view. However, if you wish to view any fields that are in the “span“ type within the form, you have to add the below code for such fields:
"showInForm": true
Example JS file (refer to specimenLabel field):

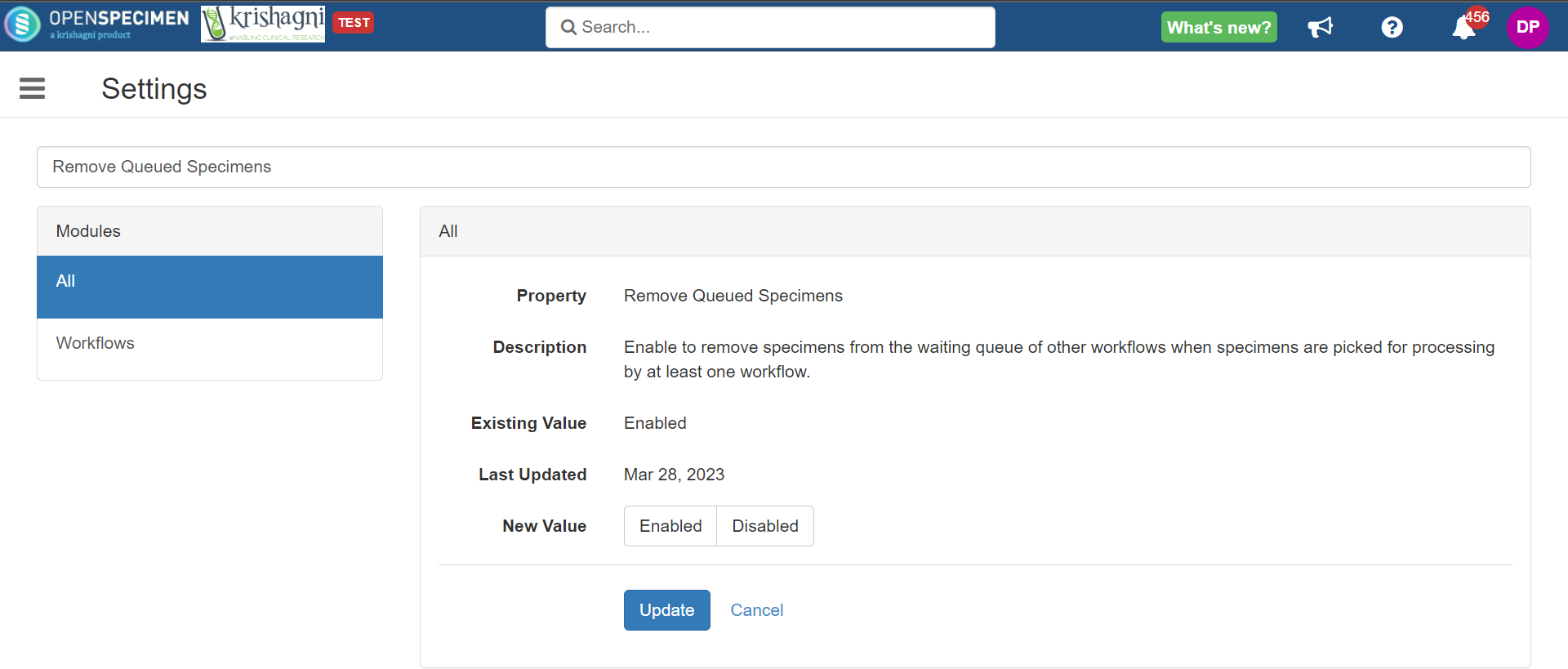
Remove Queued Specimens
If you have a similar set of rules for multiple workflows, a specimen gets added to the waiting queue multiple times for each workflow. When you pick the specimen from the waiting queue to start a new batch, you would want that specimen to be removed from the queue and no longer be used for other workflows.
Steps to enable the setting...
The specimens are removed from the waiting queue in both of the below cases:






















