Leave a comment at the end of this page or email contact@krishagni.com
Administrative screen configurations - (Auto allocation, Rule based Label Format, Lock Fields)
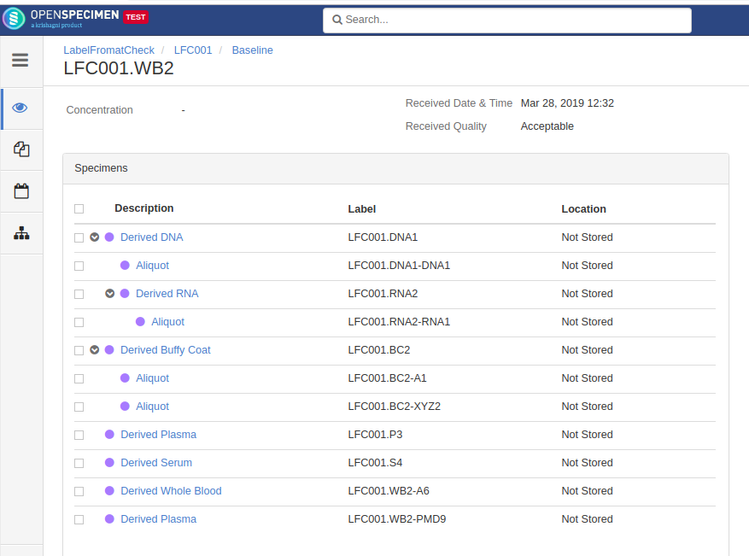
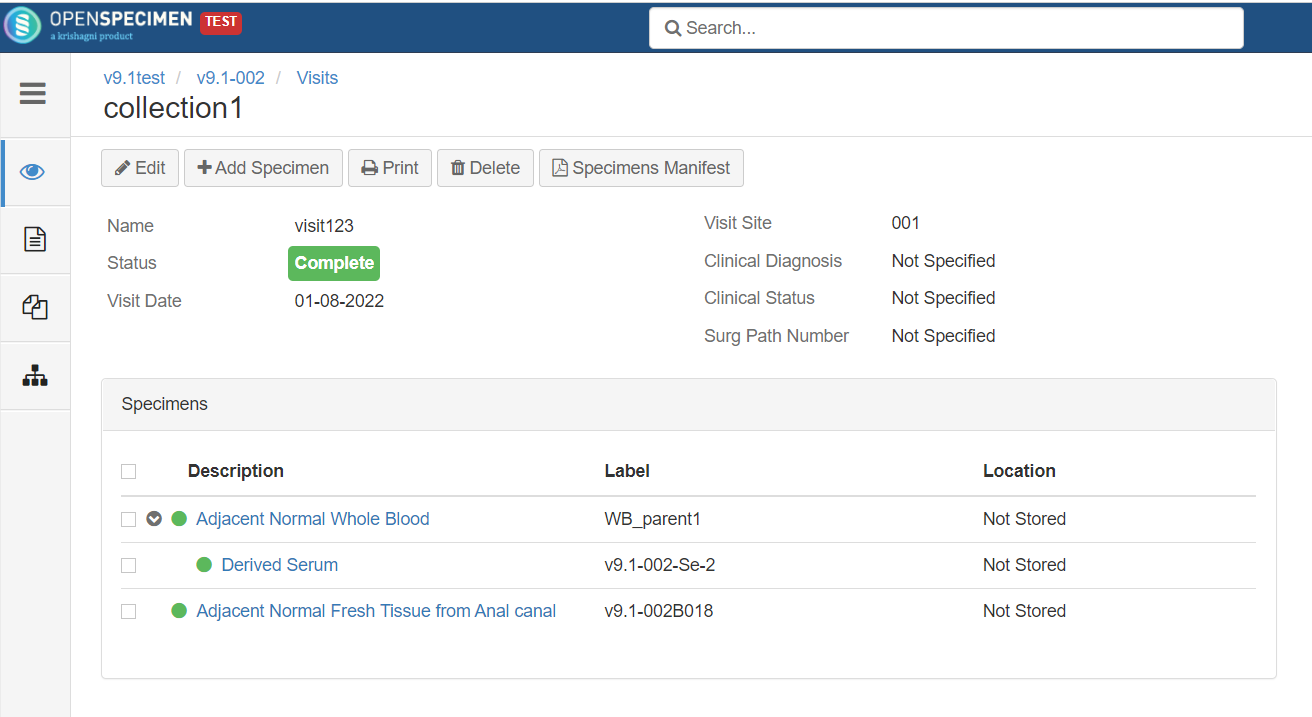
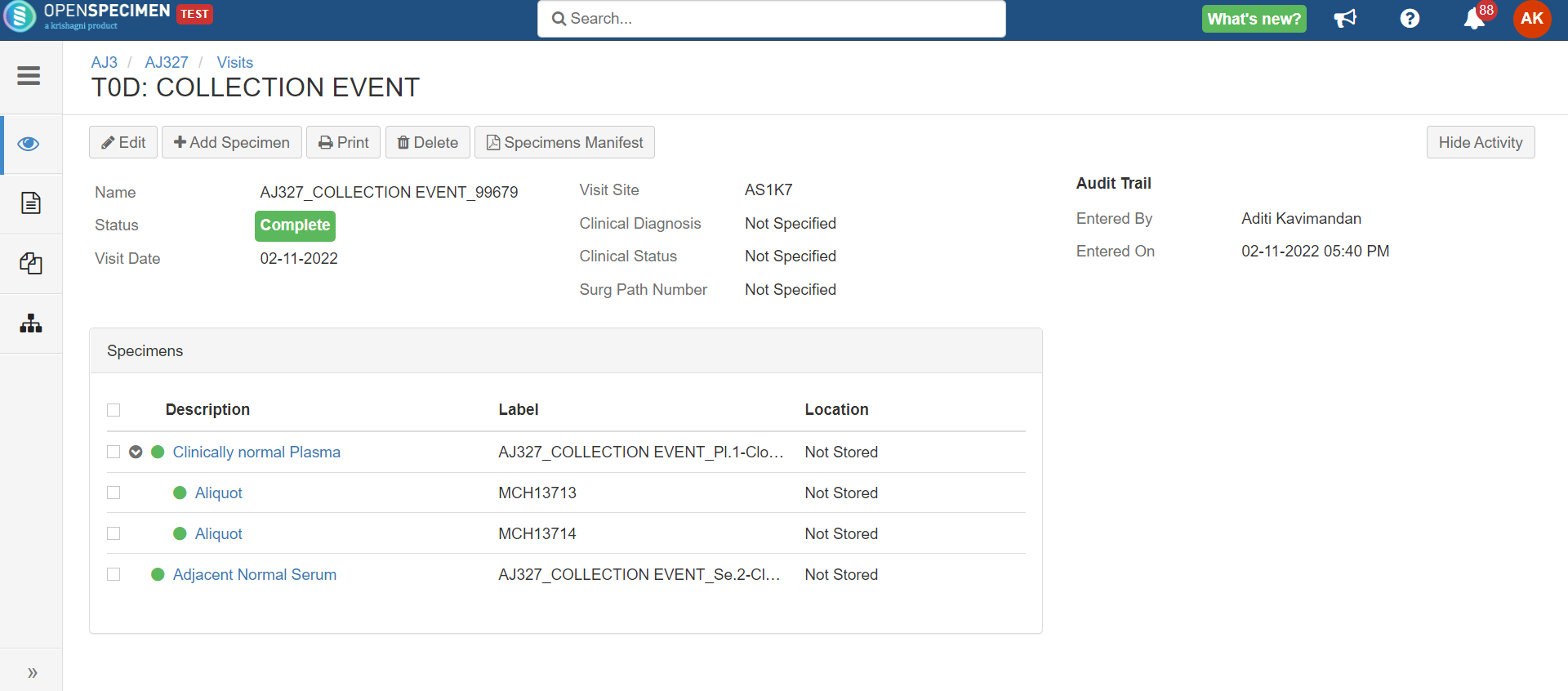
Rule-based specimen label format (v6.0)
By default, when label formats are defined in the CP, it applies to all the specimens in that CP irrespective of other conditions like specimen type. Admins can configure conditional rules to generate the specimen label formats.
Examples:
- Configure different formats for tissue specimens vs. fluid specimens
- Different format for baseline event vs. surgery event
Rules are defined in JSON. Each rule should specify the criteria that the specimen should satisfy and the specimen label format. The first matching rule is used for generating the label. Any specimen attributes, including custom attributes, can be used in the rules. The rules can be configured either at the CP or system level.
The order of label formats to use should be:
- SR level
- If the SR level is not specified, use lineage-based formats specified at the CP level.
- If lineage-based formats are not specified, use the rules-based format.
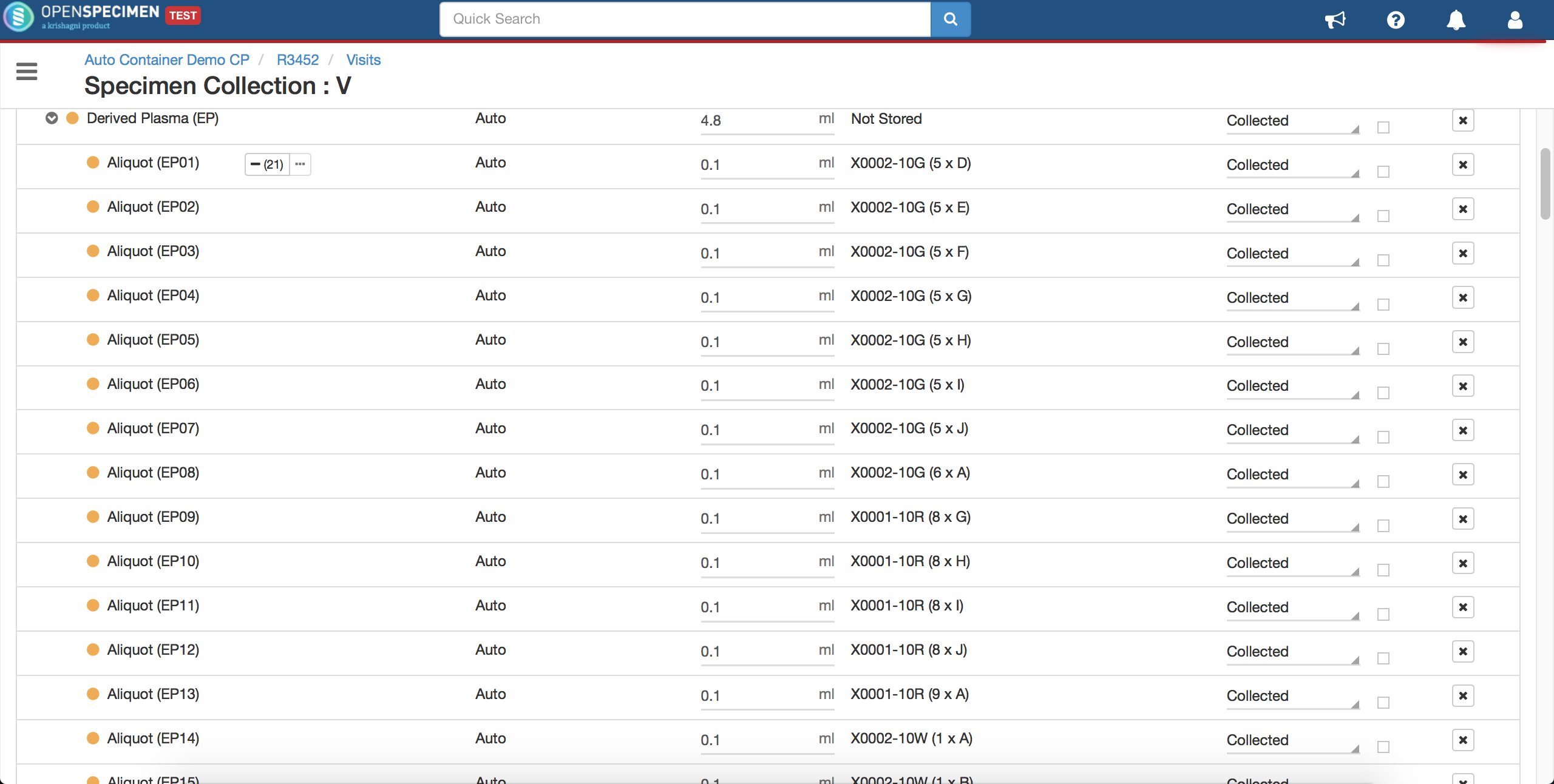
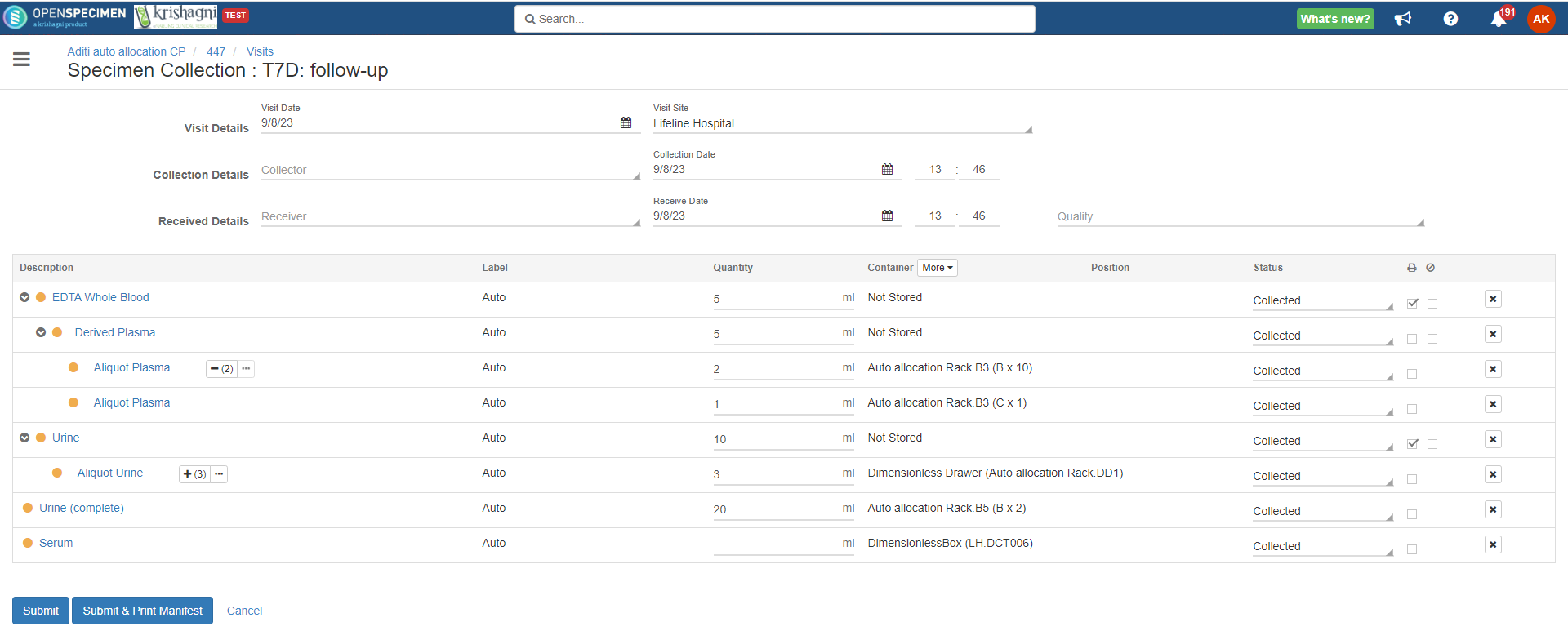
Advanced container auto-allocation
Per protocol, administrators can configure to auto-allocate containers as per different parameters. Basic settings can be achieved using the 'Collection Protocol' UI settings. Refer to 'Auto Container Allocation' a page for more details.
Complex auto allocation strategies can be set using the JSON workflow. Specimens can be auto-allocated to different storage container types during specimen collection. This can be achieved by setting up criteria in various fields, as listed below.
From V10.3 it's possible to include dimensionless container types for condition-based auto allocation. Refer to wiki page to create dimensionless container type.
| Level | Field | Variable to be used |
|---|---|---|
| Visit | Event label | specimen.eventLabel |
| Specimen | Quantity | specimen.initialQty |
| Specimen | Type | specimen.type |
| Specimen | Lineage | specimen.lineage |
| Specimen | Requirement code | specimen.reqCode |
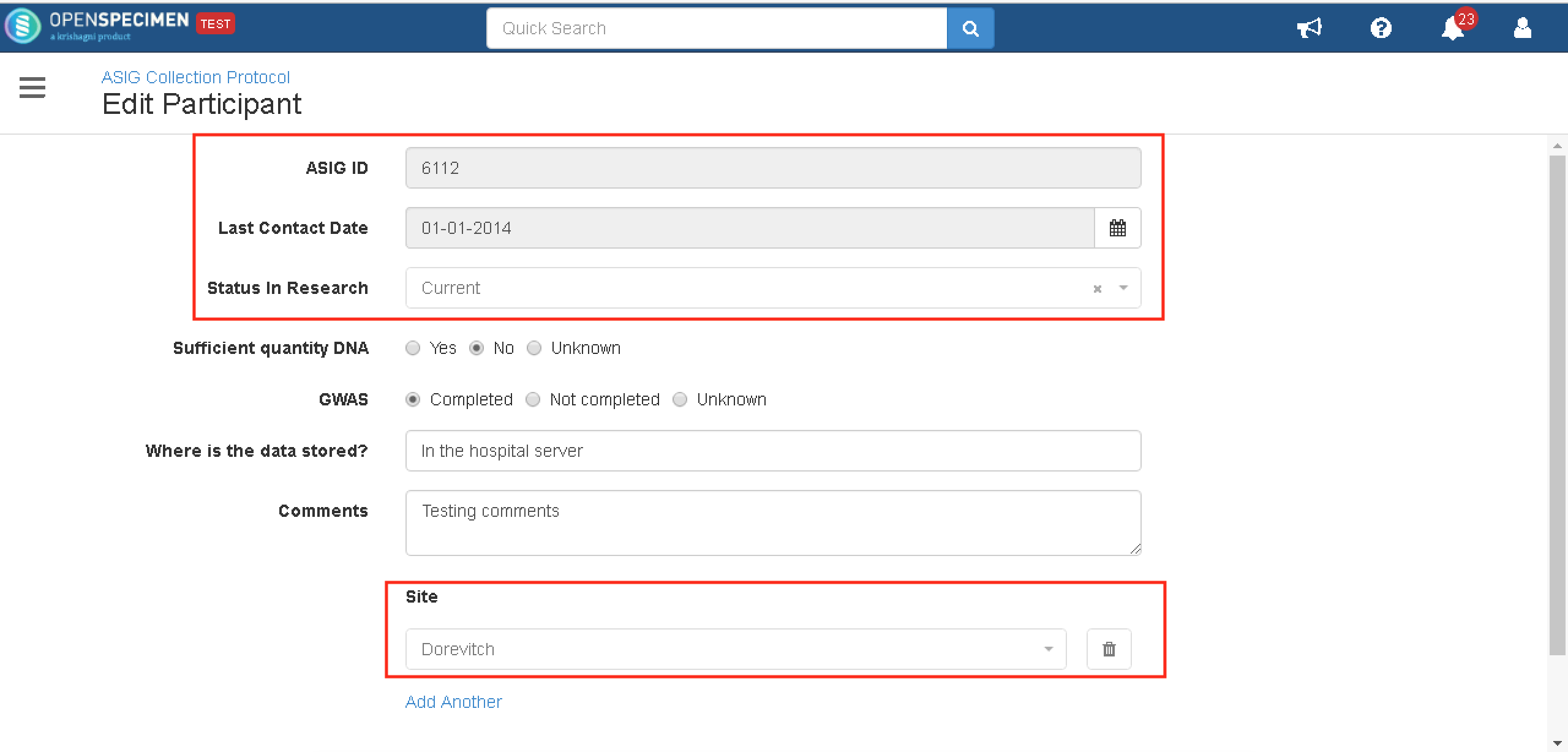
Lock fields at the participant level
Using SDE, you can lock the participant fields on the Openspecimen UI. These fields are populated from an external source, due to which biobanks want these fields to be non-editable.
The source of the fields is the name of the DB from where the participant data is being populated. In the example below, the source is 'ASIG'. In the OpenSpecimen database table catissue_participant, the source column for the participants should be 'ASIG' so that the fields will be locked.
This JSON should be set at the system level since the participants are registered across CPs.
Once the JSON is configured, the participant edit page will be displayed as the screenshot below.
Leave a comment at the end of this page or email contact@krishagni.com