...
Attribute | Description |
"type" | Type of the task |
"name" | Name of the task. This must be unique within the workflow |
"title" | Title of the task. This will be displayed on the UI. |
"shortDescription" | Short description of the task that needs to be displayed along with the title. |
"activityStatus" |
|
"longDescription" | Detailed description of the task and steps to follow. |
"jsonConfig" | Indicates if the task is configured within the JSON file or not.
|
"starterTask" | Indicates if this is the first task in workflow or not.
|
"optional" | Indicates if this task is mandatory or not.
|
"onSkipAction" | Adds a skip button to the task. Using this button, you can skip the specific task by mentioning the reason for the deviation.
|
"inputs" | Specify the input for each task. This needs to be configured for all tasks except the starter task. For example, the input of the second task will be the first task. The input for the third task will be the second task, and so on. |
"userGroups" | You need to specify the user groups who would be using this workflow. By default, only super admins have access to all the workflows within the system. To share it with user groups, you need to specify it in the workflow. |
...
Type | Description |
|---|---|
update-specimens | Any task that updates the existing default or custom fields field’s value of a specimen are is considered as an update specimens task. Example: Updating received quality, storage location, specimen quantity, etc. |
create-pooled | Allows you to create pooled specimens |
form-data-entry | Allows you to add specimen events or custom form fields |
create-derivatives | Create derivative task |
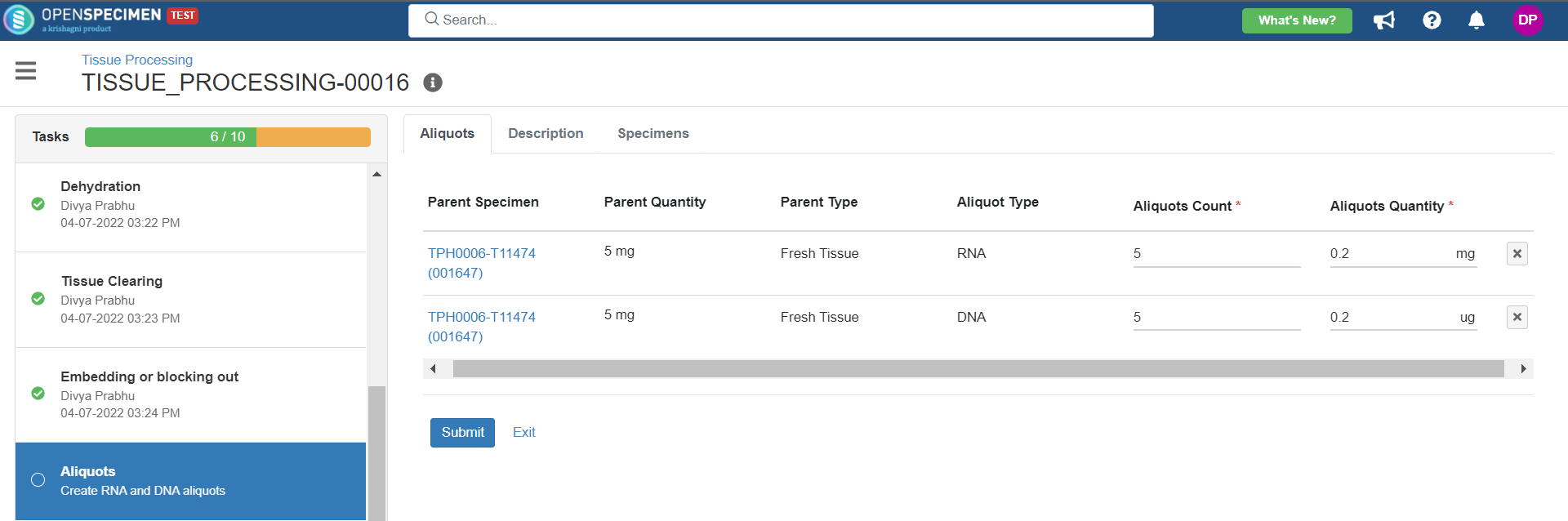
create-aliquots | Create aliquots task |
group-assigner | Allows you to add an assigner where you can select the next steps for the specimens |
...
| Expand | ||
|---|---|---|
| ||
|
Examples
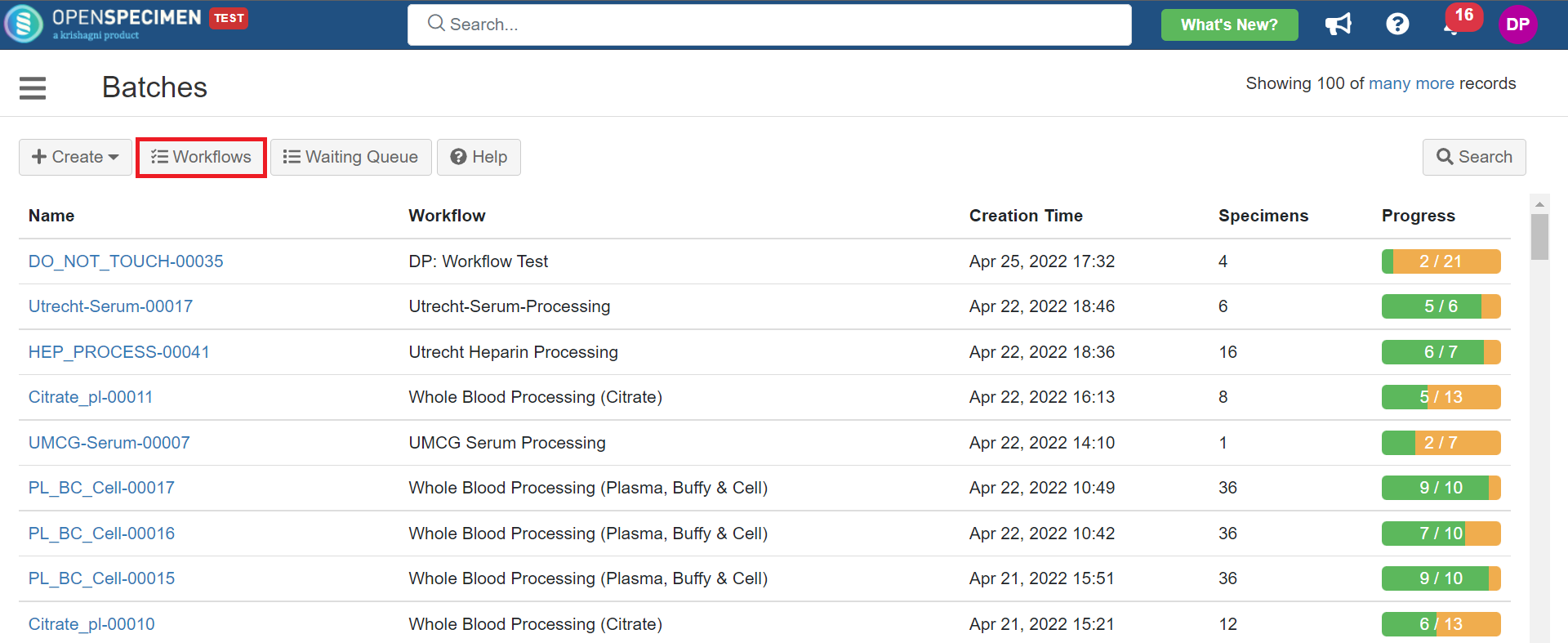
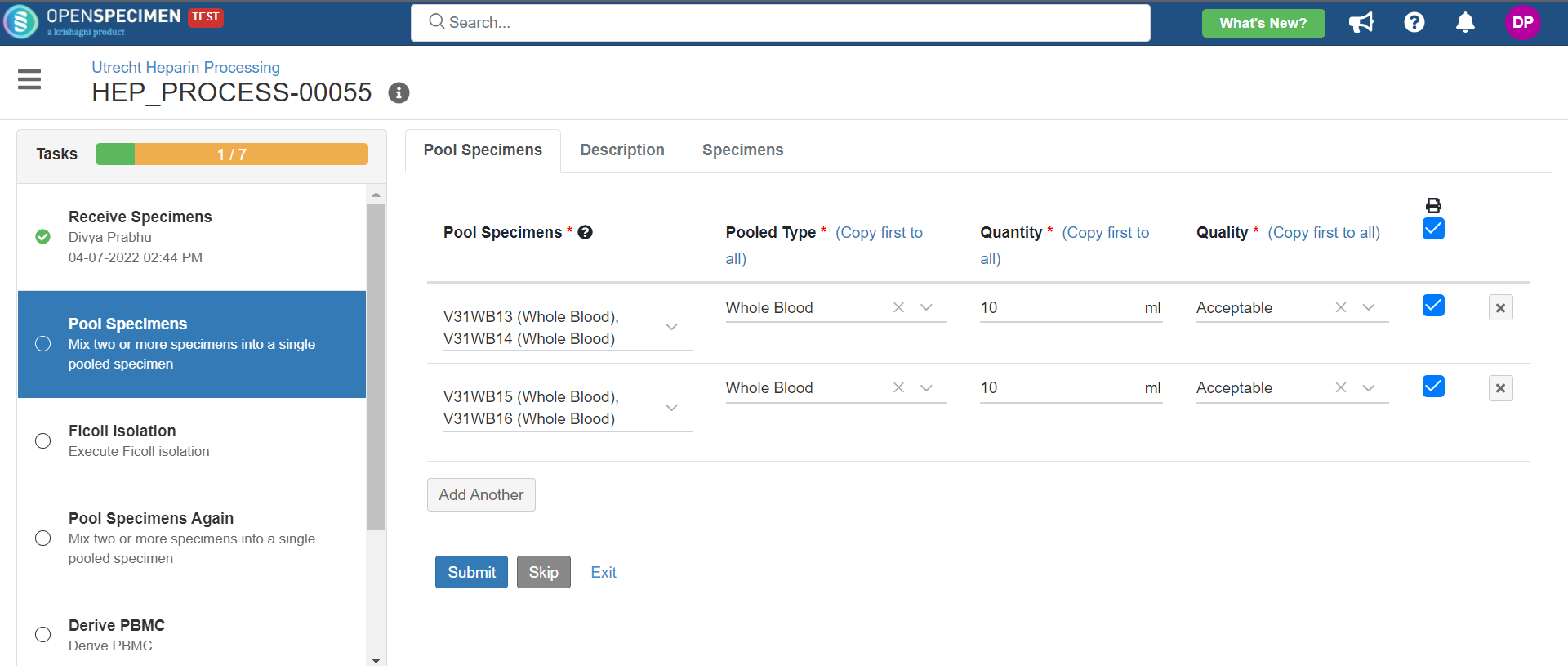
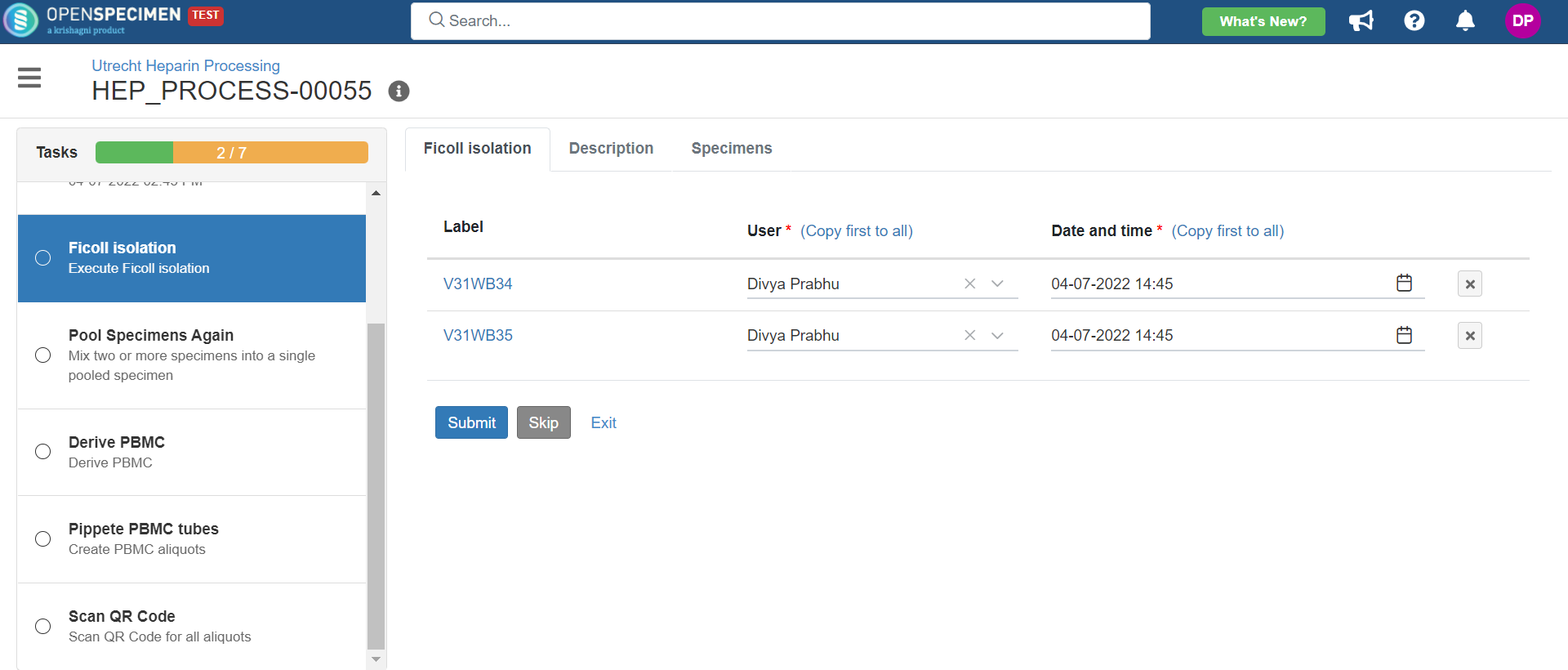
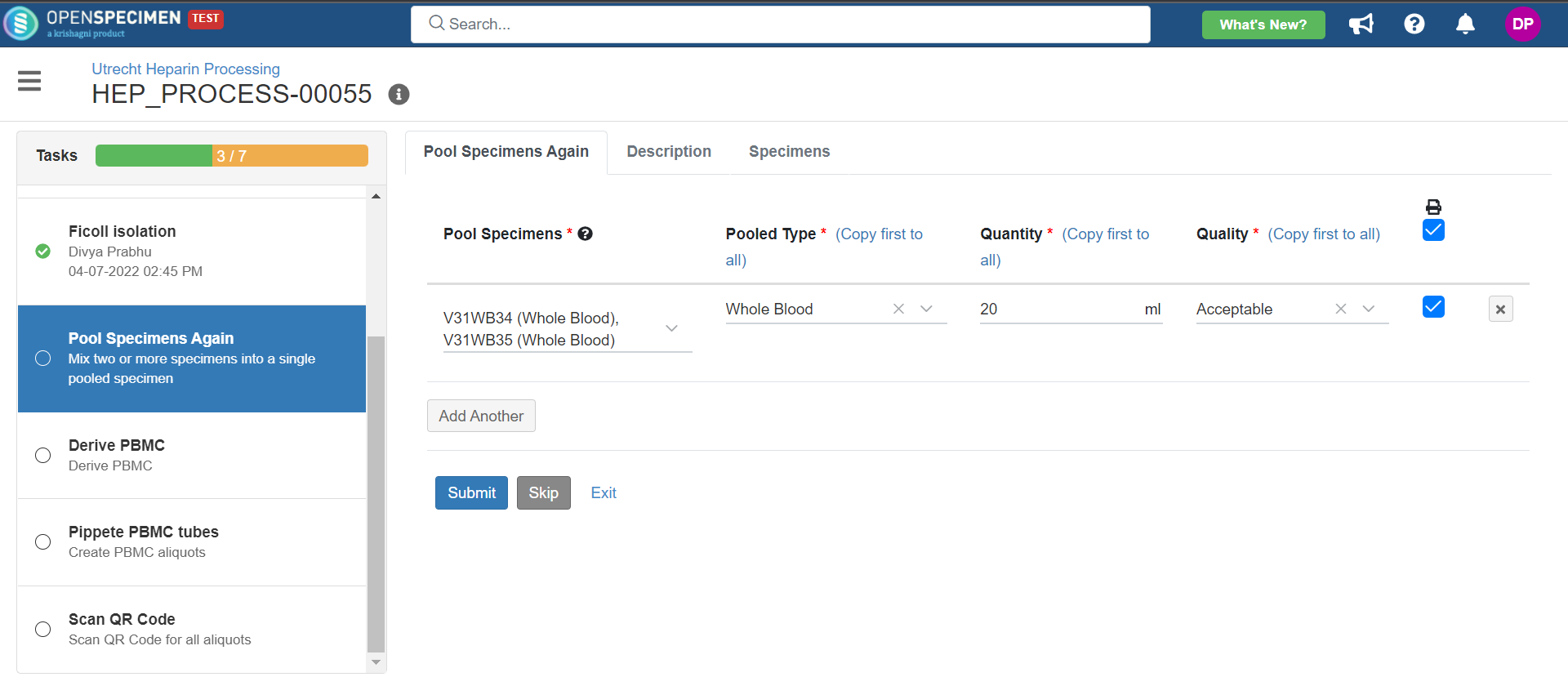
1. Heparin Processing
This example is for whole blood specimens collected in a heparin tube and further processing.
| Expand | ||
|---|---|---|
| ||
Below is the zip file for the heparin processing workflow: View file | | name | hep-process
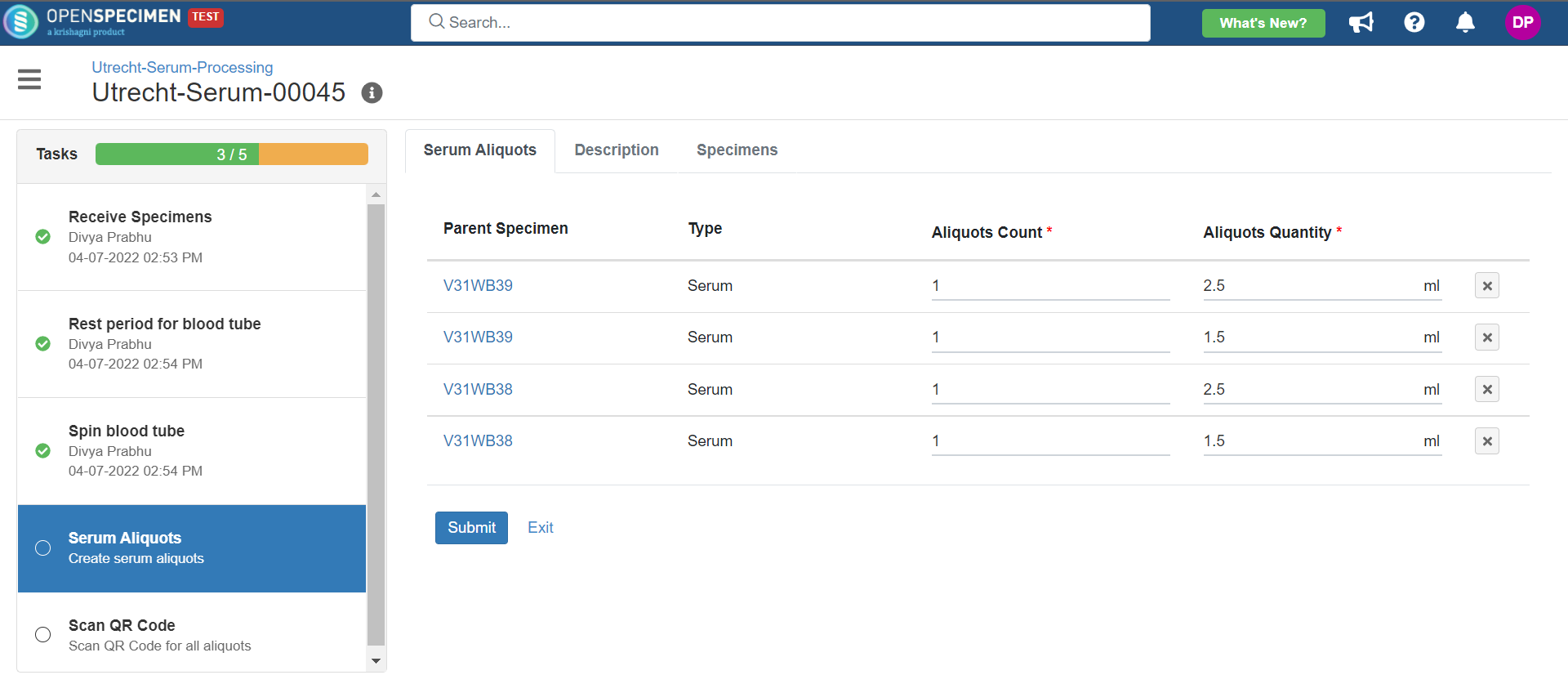
2. Serum Processing
This example is for whole blood specimens collected in a serum separator vacutainer and further processing.
| Expand | ||||
|---|---|---|---|---|
| ||||
Below is the zip file for the serum processing workflow:
|
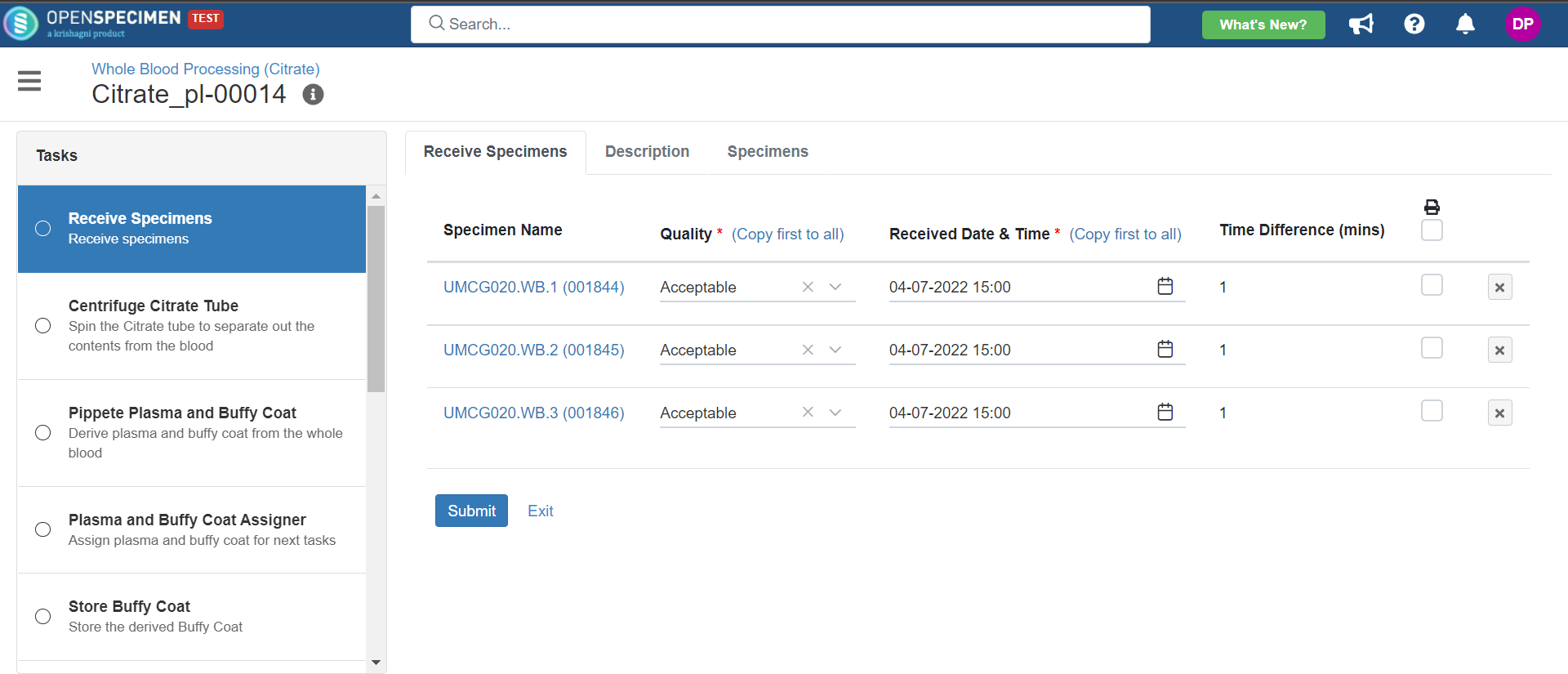
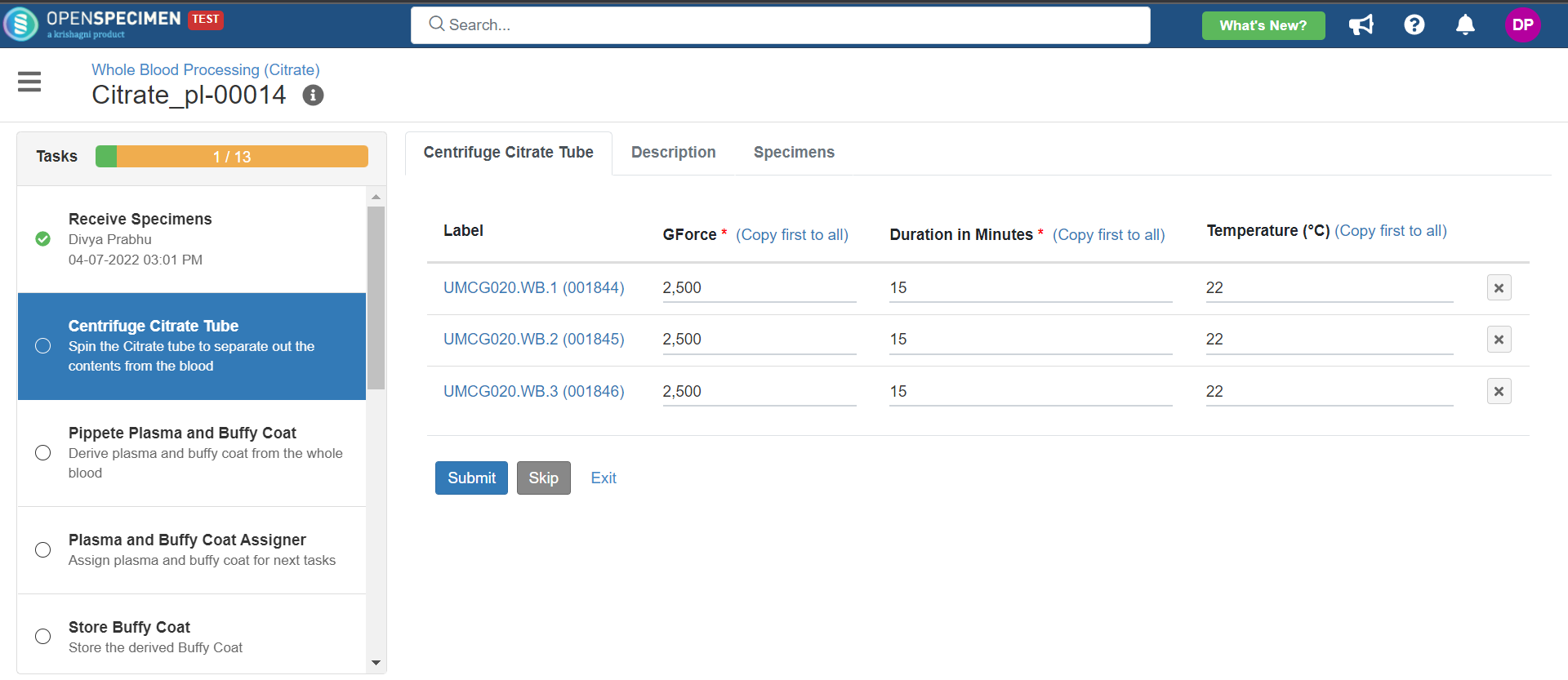
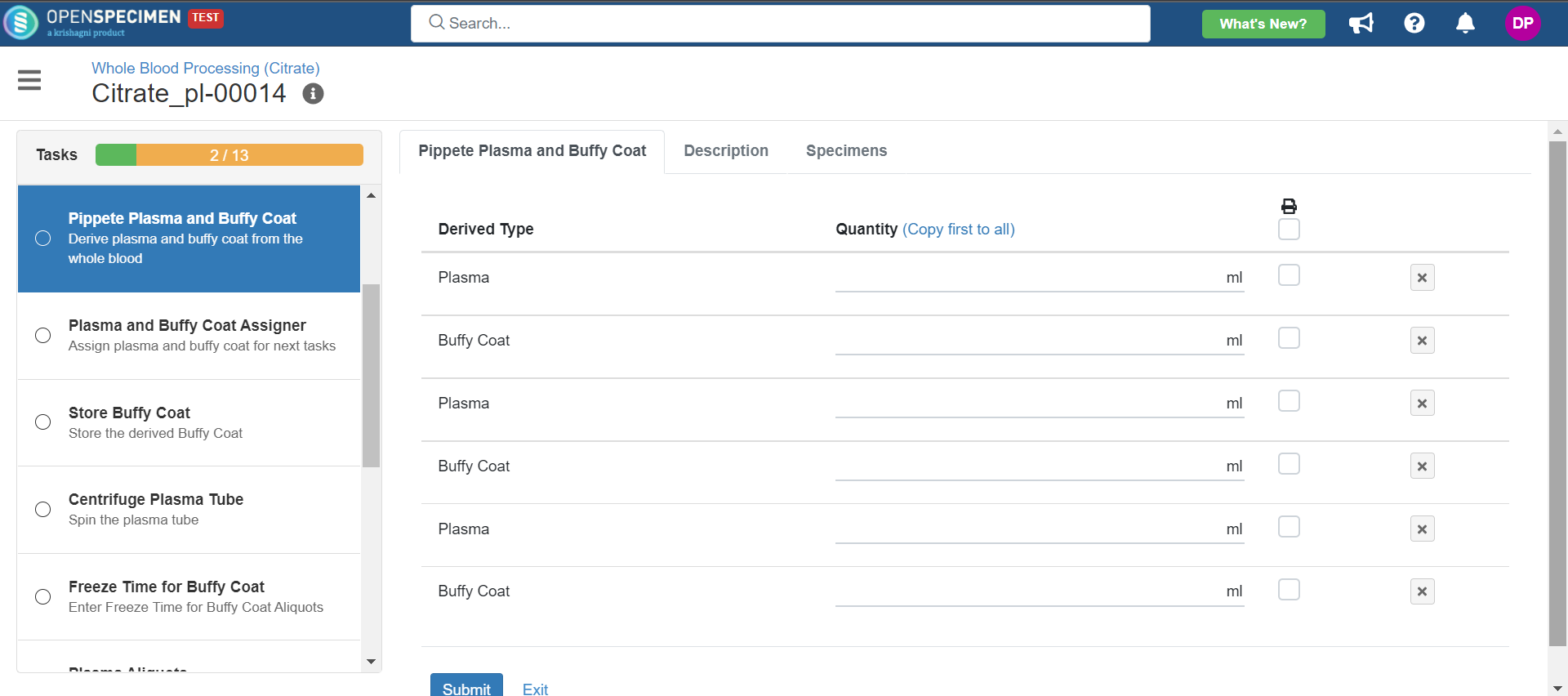
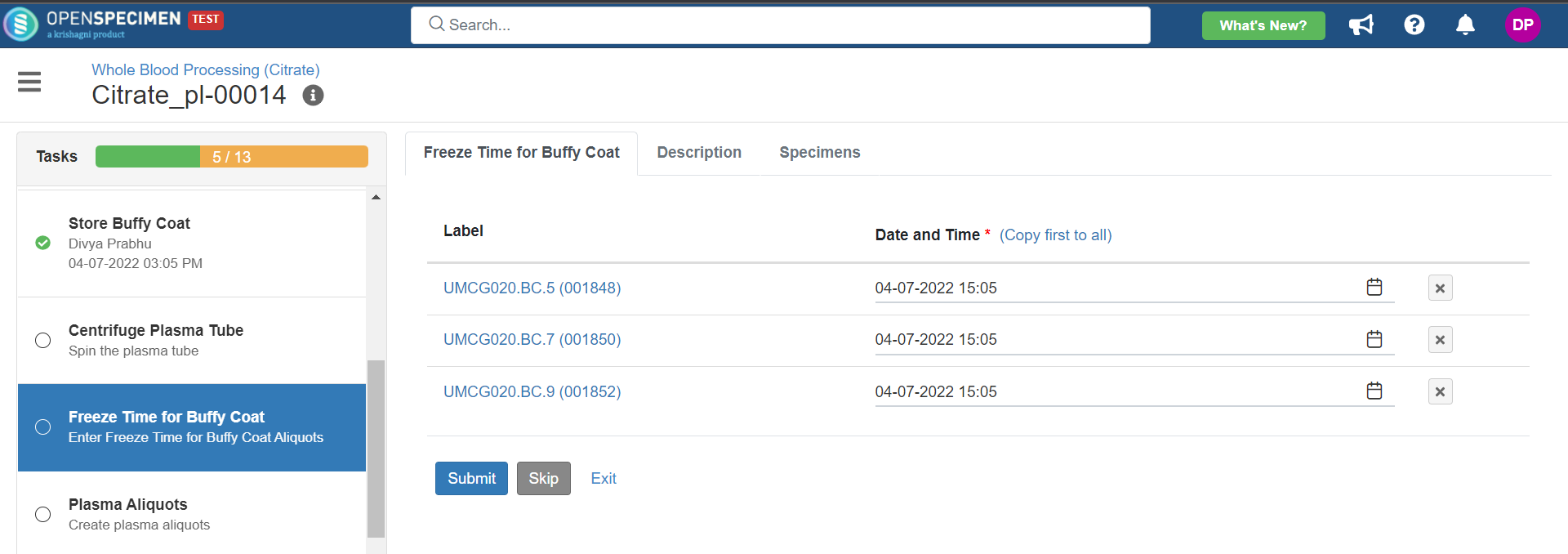
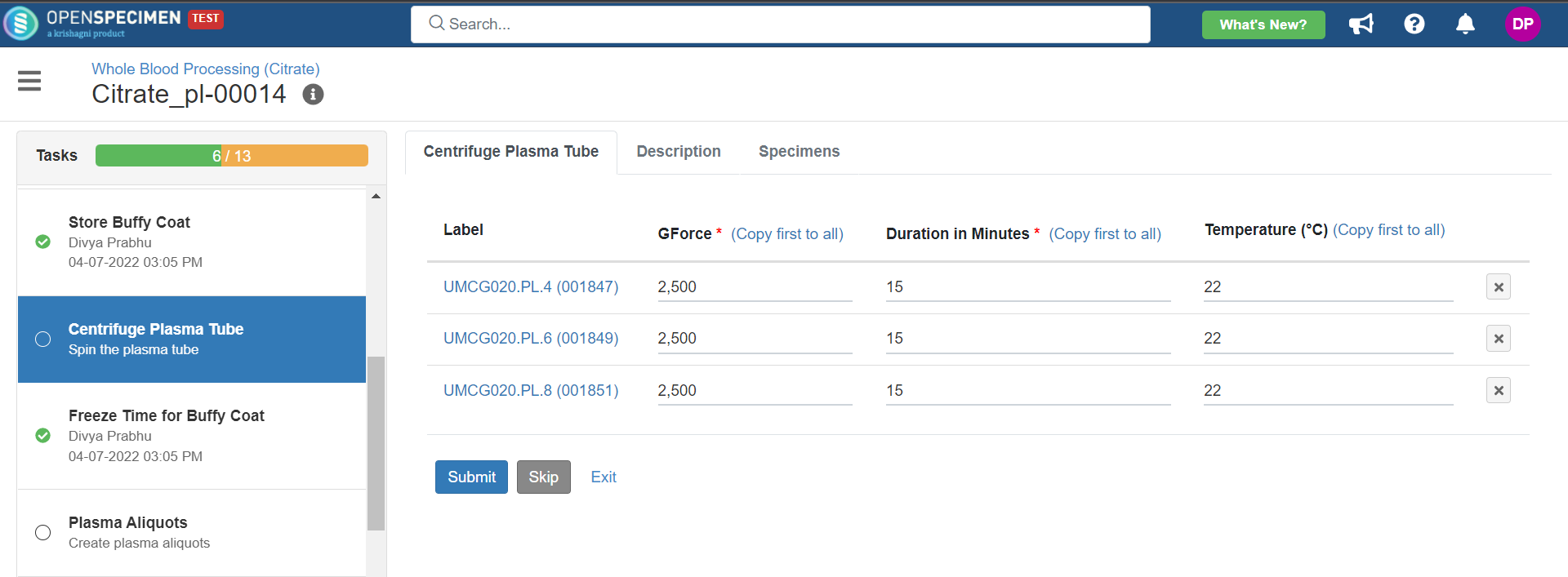
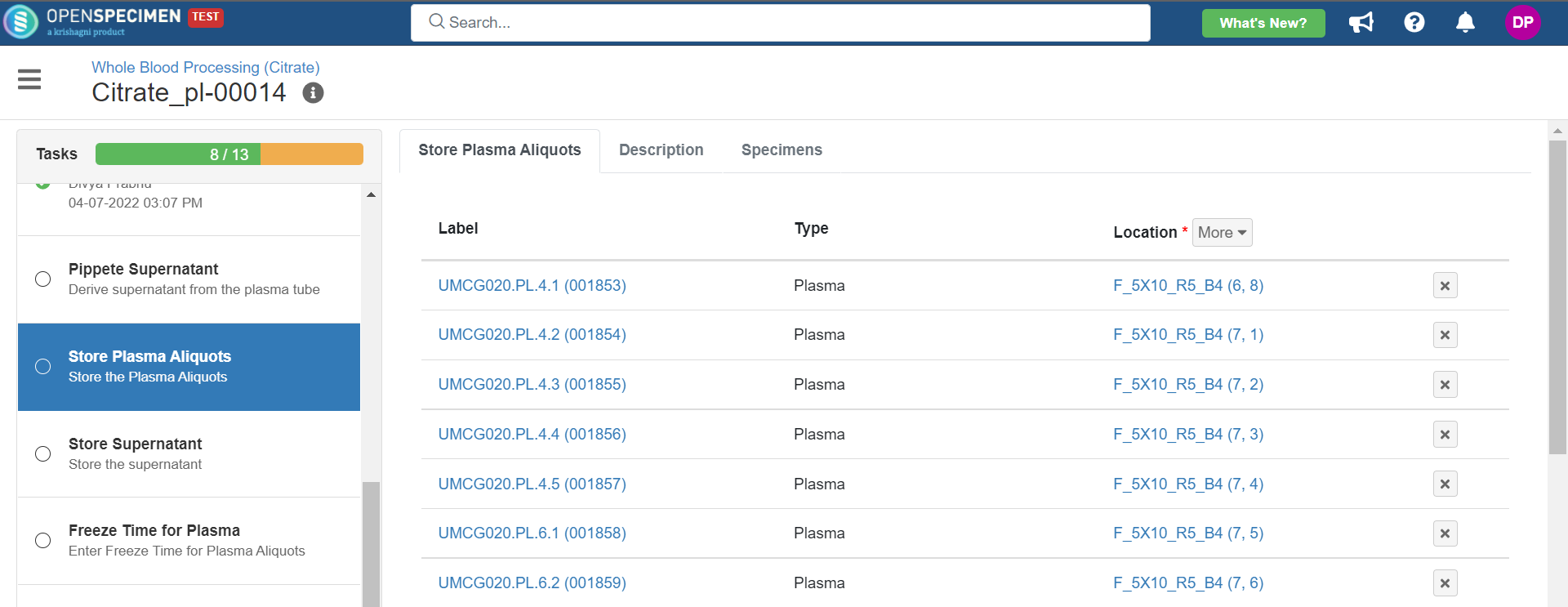
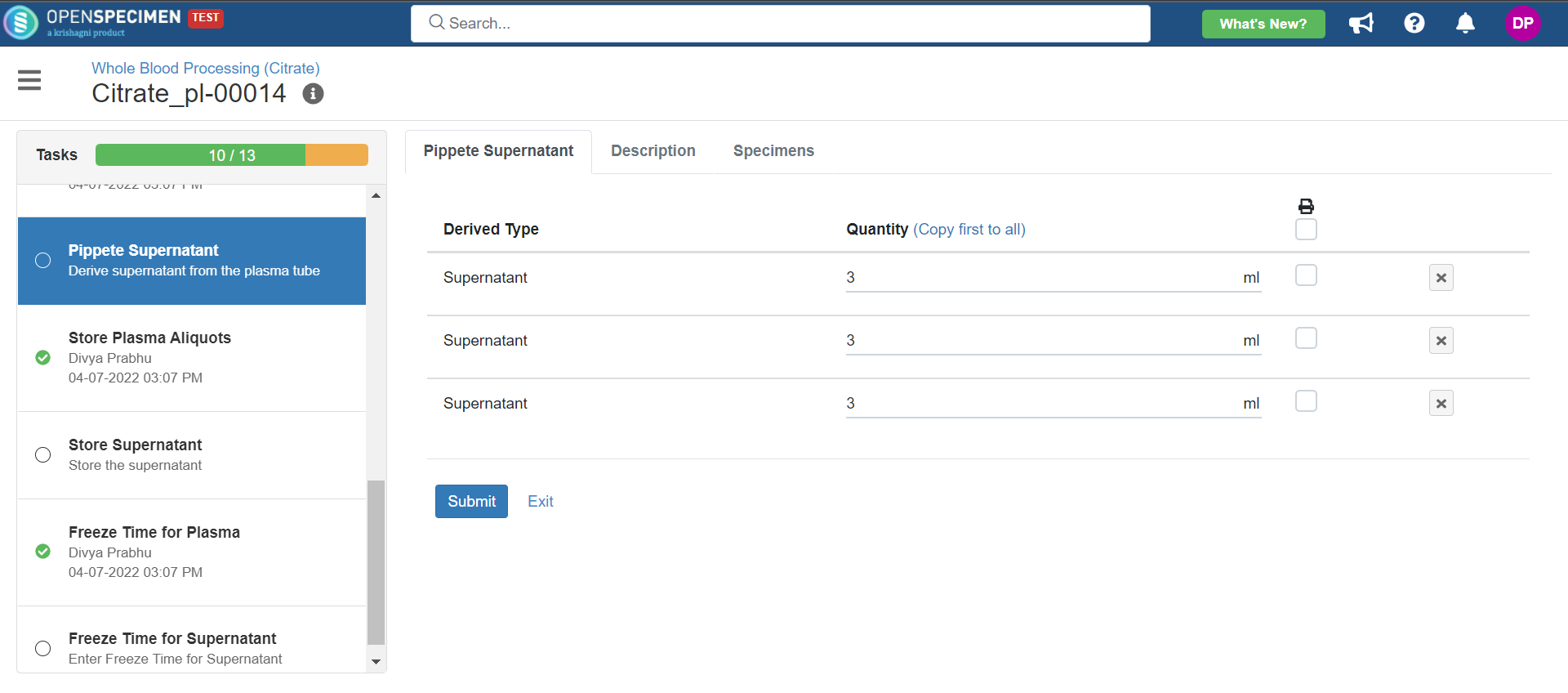
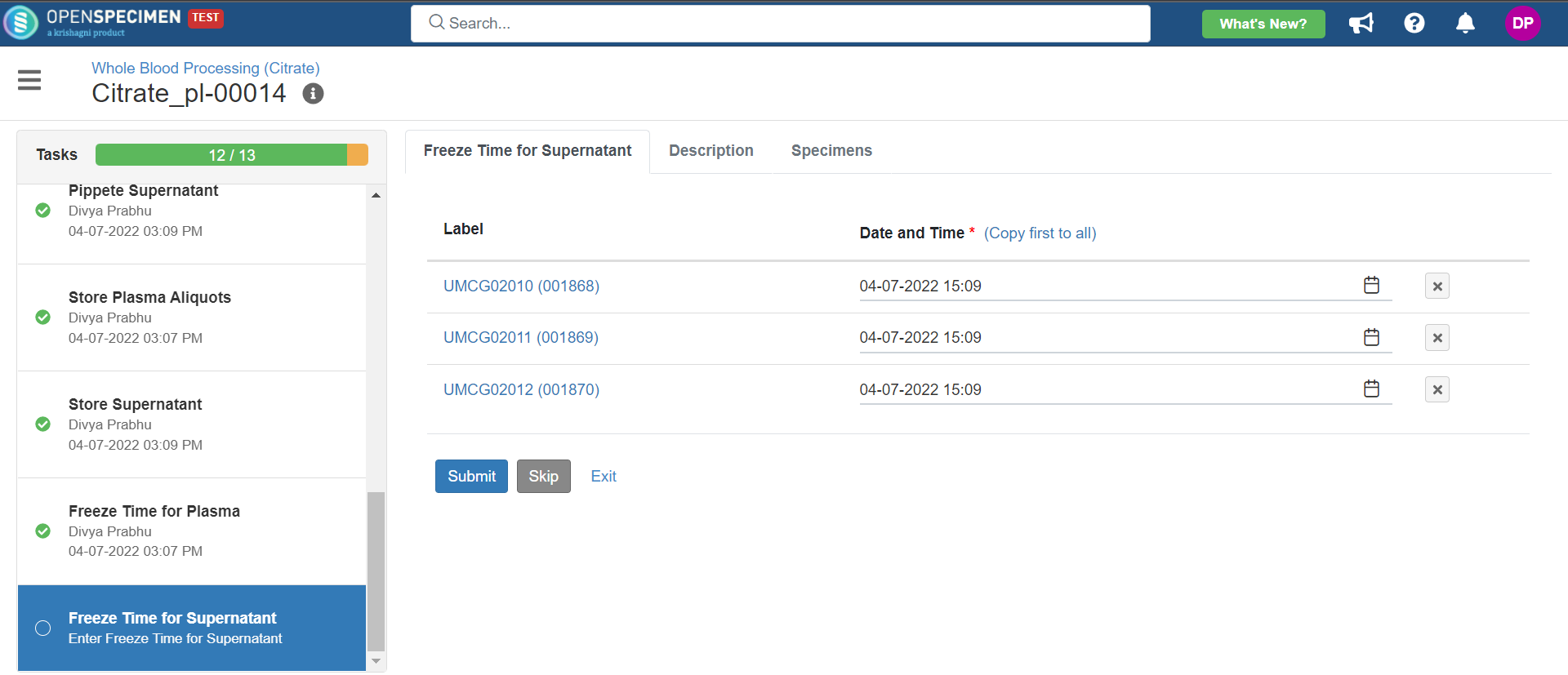
3. Whole Blood Processing in Citrate Tube
This example is for whole blood specimens collected in a citrate tube and further processing.
| Expand | ||||
|---|---|---|---|---|
| ||||
Below is the file for the whole blood processing in the citrate tube workflow:
|
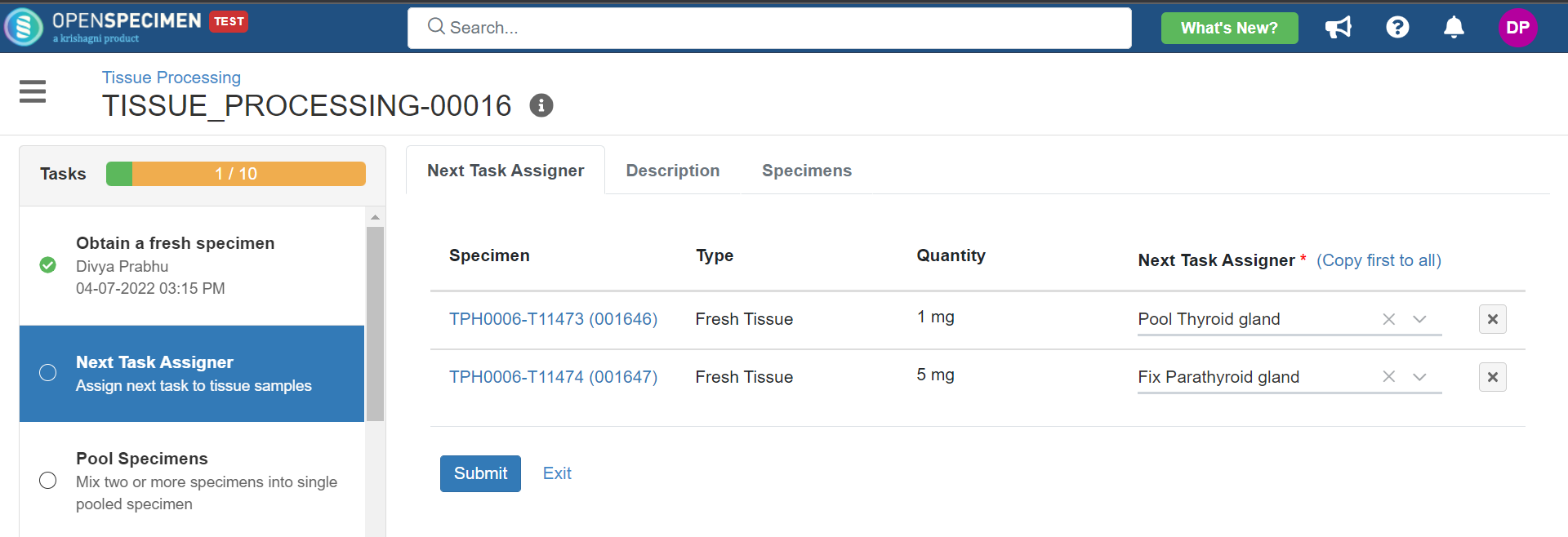
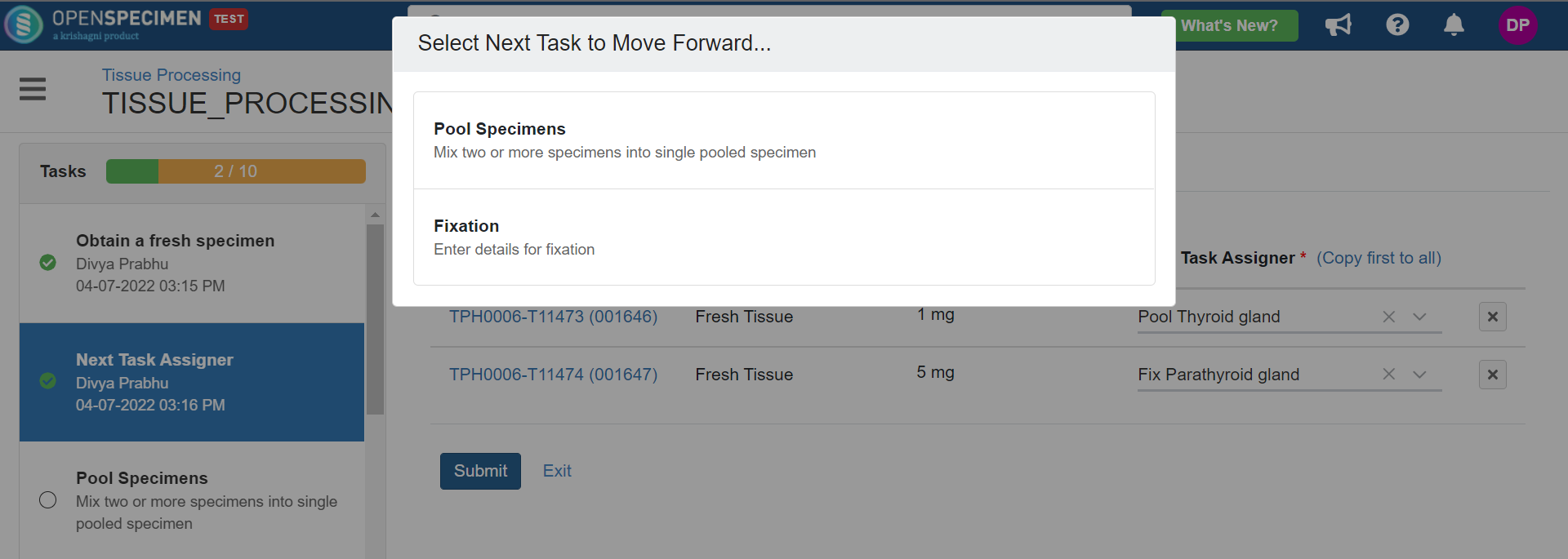
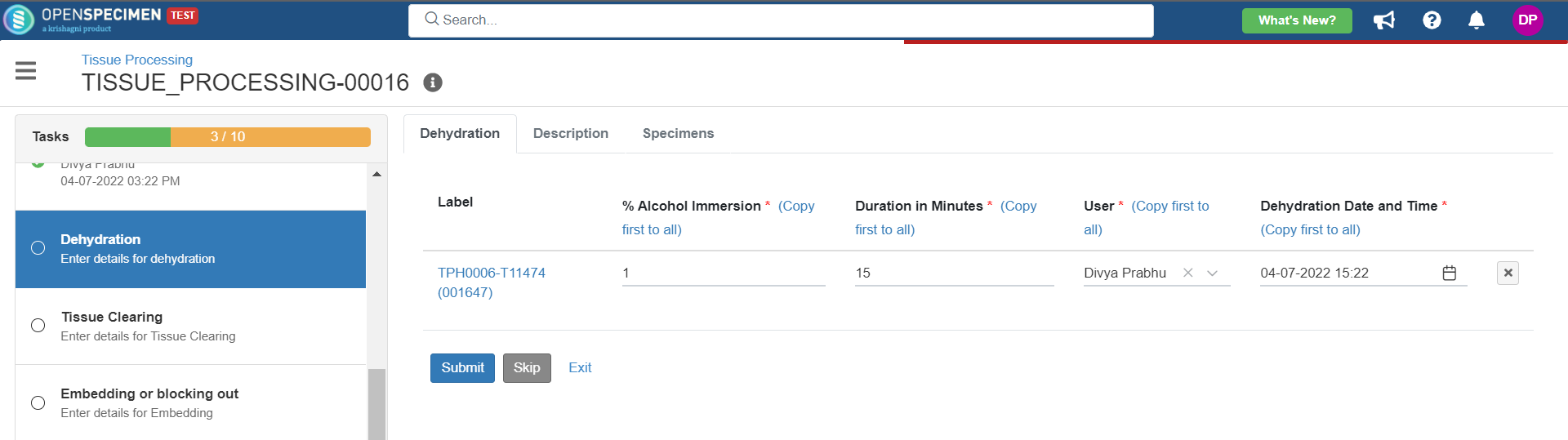
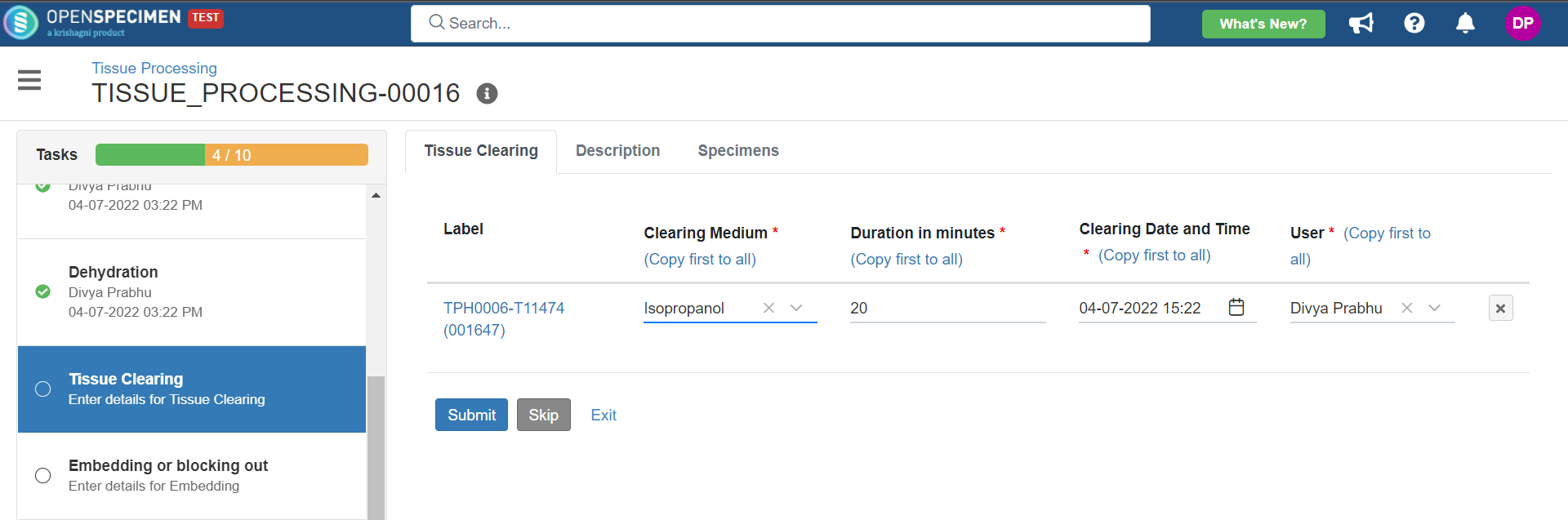
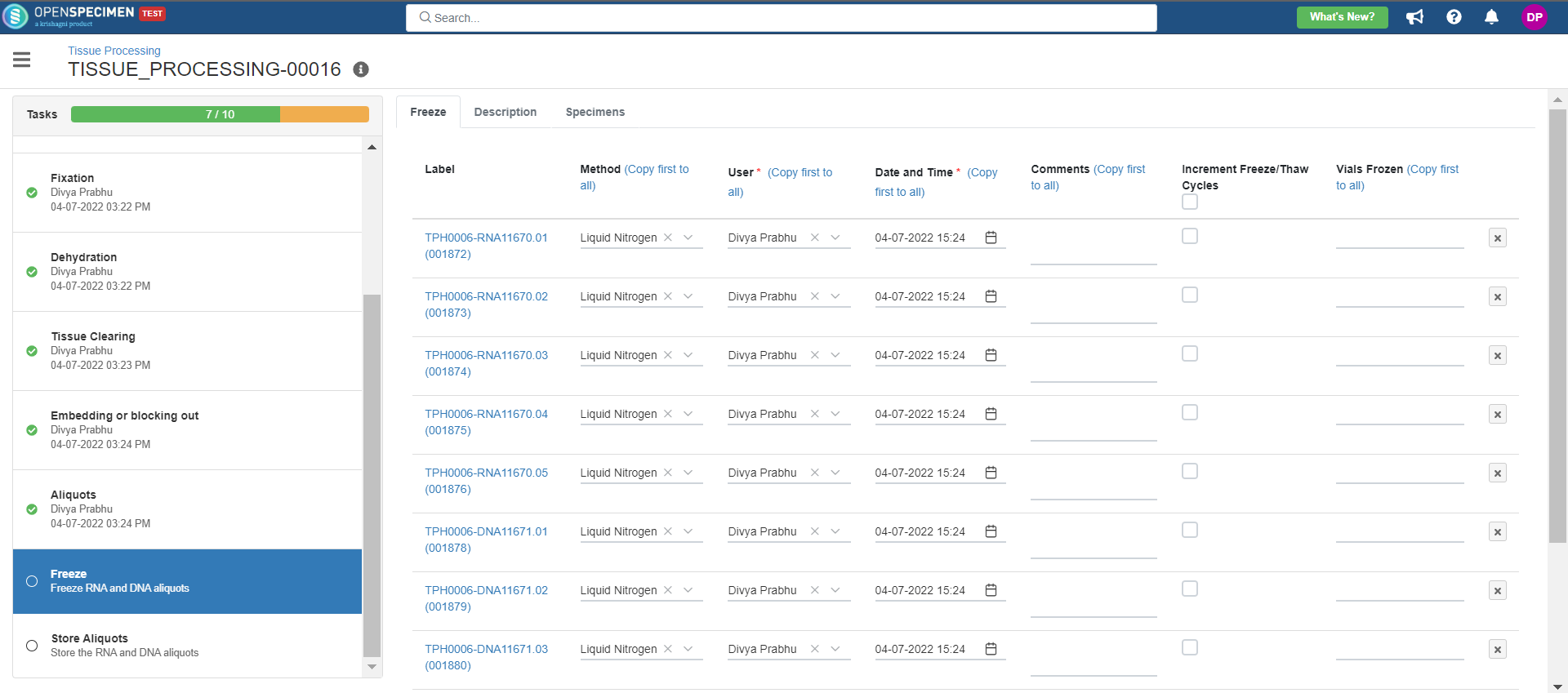
4. Tissue Processing
This example is for tissue specimen processing.
| Expand | ||
|---|---|---|
| ||
Below is the file for the whole blood processing in the citrate tube workflow: View file | | name | tissue-processing.zip